Adding statistical significance indicators to your data visualizations is crucial for communicating research findings effectively. When working with ggplot2 in R, displaying p-values directly on scatter plots helps viewers immediately understand the statistical relationship between variables.
In this comprehensive tutorial, you’ll learn how to annotate ggplot2 plots with p-values from linear regression analysis. We’ll cover two powerful methods: using the standard geom_text() function and the enhanced geom_richtext() from the ggtext package for customized annotations
Required R packages: tidyverse, palmerpenguins, ggtext, glue, and broom for statistical modeling.
What You’ll Learn
- How to add p-values to ggplot2 scatter plots
- Performing linear regression analysis and extracting statistical results
- Using geom_text() for basic p-value annotations
- Customizing p-value displays with geom_richtext() and styling options
- Best practices for statistical significance visualization in R
library(tidyverse) library(palmerpenguins) library(ggtext) library(glue) theme_set(theme_bw(16))
We will use Palmer penguin dataset to make the scatter plot.
penguins |> head() # A tibble: 6 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 # ℹ 2 more variables: sex <fct>, year <int>
How to Add P-value to a plot in ggplot2
Here is a scatter plot between two numerical variables from penguins dataset.
p1 <- penguins |>
ggplot(aes(flipper_length_mm, body_mass_g))+
geom_point(aes(color=species))+
geom_smooth(method = "lm", formula = y ~ x) +
theme(legend.position = "none")+
labs(title="How to annotate the plot with p-value")
p1
ggsave("how_to_annotate_with_p_value_ggplot2.png")
In order to annotate the plot with p-value, let us first perform the statistical test using linear regression model and save p-value in a dataframe. In addition to the p-value, we also create variables to help annotate the plot with the p-value.
Do statistical test and save result in dataframe
In the code below, we use tidyverse framework to perform linear regression and store the results in a dataframe.
pval_df <- penguins |>
summarize(lm_mod = list(lm(flipper_length_mm~ body_mass_g)),
lm_res = map(lm_mod, broom::tidy)) |>
unnest(lm_res) |>
filter(term=="body_mass_g") |>
select(p.value) |>
mutate(flipper_length_mm=200,
body_mass_g=6000,
label=glue("p-val: {signif(p.value,3)}"))
This is how the dataframe with p-value looks like this.
pval_df
# A tibble: 1 × 4
p.value flipper_length_mm body_mass_g label
<dbl> <dbl> <dbl> <glue>
1 4.37e-107 200 6000 p-val: 4.37e-107
Adding statistical significance as annotation with geom_text()
Now we can use geom_text() function available in ggplot2 to add the p-value as annotation to the plot.
p1 +
geom_text(
data = pval_df,
aes(label = label),
hjust = 1, vjust = 1,
size=6
)+
labs(title="Annotating a plot with p-value")
ggsave("annotate_plot_with_p_value_ggplot2.png")
Annotating a plot with p-value using ggtext’s geom_richtext()
We can further customize the annotation using geom_richtext() function from ggtext package to add color and a box around the annotation text.
p1+
geom_richtext(
data = pval_df,
aes(
label = label#,
#fill = after_scale(alpha(colour, .2))
),
text.colour = "purple",
hjust = 1, vjust = 1,
size=6
)+
labs(title="Annotating a plot with p-value")
ggsave("annotate_plot_with_p_value_ggplot2_example2.png")
Explore the Complete ggplot2 Guide
35+ tutorials with code: scatterplots, boxplots, themes, annotations, facets, and more—tested and beginner-friendly.
Visit the ggplot2 Hub → No fluff—just code and visuals.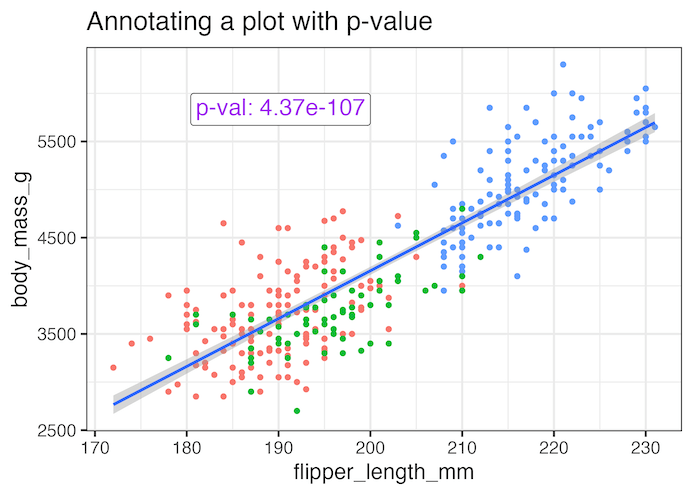
1 comment
Comments are closed.