In this tutorial, we will learn how to add p-value to each facet plot made with facet_wrap() function in ggplot2. We will use scatter plot example, where we have multiple scatter plots using facet_wrap and we have done linear regression analysis to find the statisitical significance of the association.
Here we will show with how to annotate the scatter plots with p-value showing the statistical significance of the association with two examples. We will use geom_text() & geom_richtext() functions in combination with packages glue and ggtext.
library(tidyverse) library(palmerpenguins) library(ggtext) library(glue) theme_set(theme_bw(16))
We will use Palmer penguin dataset to make the scatter plot.
penguins |> head() # A tibble: 6 × 8 species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torgersen 39.1 18.7 181 3750 2 Adelie Torgersen 39.5 17.4 186 3800 3 Adelie Torgersen 40.3 18 195 3250 4 Adelie Torgersen NA NA NA NA 5 Adelie Torgersen 36.7 19.3 193 3450 6 Adelie Torgersen 39.3 20.6 190 3650 # ℹ 2 more variables: sex <fct>, year <int>
How to annotate multiple plots with p-values
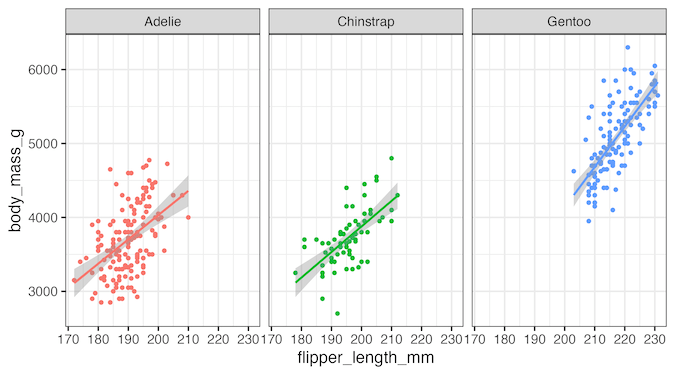
In this post, we will learn how to add p-value to multiple plots made with facet_wrap() in ggplot2. Let us make scatter plots using facet_wrap().
p1 <- penguins |>
ggplot(aes(flipper_length_mm, body_mass_g, color=species))+
geom_point()+
geom_smooth(method = "lm", formula = y ~ x) +
facet_wrap(~species)+
theme(legend.position = "none")
p1
ggsave("how_to_annotate_multiple_plots_with_p_value_ggplot2.png",width=9, height=5)
To add p-value as annotation to each facet, let us first perform linear regression analysis using tidyverse framework and save p-value in a data frame. To do this we use broom packages’ tidy() function.
Since we will be adding p-value as annotation to each facet, we need x & y co-ordinates for placing the p-values on the plot. We add the coordinates in the scale of the variables we are plottting.
pval_df <- penguins %>%
group_by(species) %>%
summarize(lm_mod = list(lm(body_mass_g~flipper_length_mm) ),
lm_res = map(lm_mod, broom::tidy)) |>
unnest(lm_res) |>
filter(term=="flipper_length_mm") |>
select(species, p.value) |>
mutate(
body_mass_g= rep(6100,3), flipper_length_mm= c(220,220,220),
label=glue("p_val = {signif(p.value,digits=3)}"))
And this is how our dataframe containing p-value and the coordinates to annotate the plots with p-values look like.
pval_df # A tibble: 3 × 5 species p.value body_mass_g flipper_length_mm label <fct> <dbl> <dbl> <dbl> <glue> 1 Adelie 1.34e- 9 6100 220 p_val = 1.34e-09 2 Chinstrap 3.75e- 9 6100 220 p_val = 3.75e-09 3 Gentoo 1.33e-19 6100 220 p_val = 1.33e-19
Annotating each facet with p-value using ggtext’s geom_richtext()
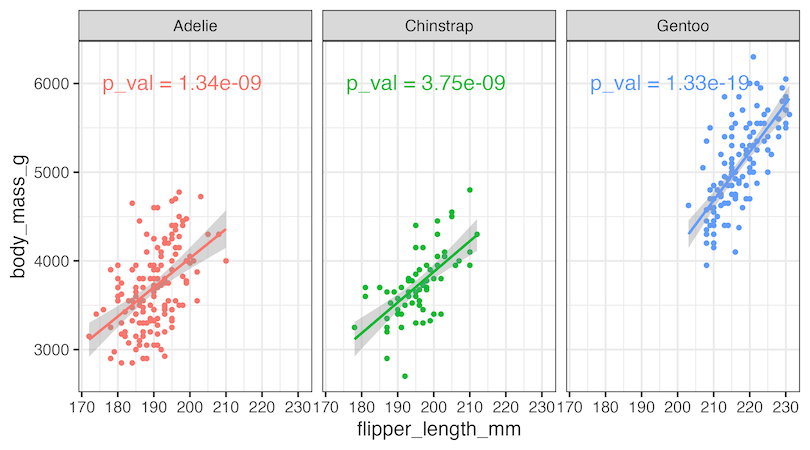
We can now use geom_text() function to add p-value as a label to the plot as shown below.
p1+
geom_text(
data = pval_df,
aes(label = label),
hjust = 1, vjust = 1,
size=6
)
ggsave("annotate_multiple_plots_with_p_value_ggplot2.png", width=9, height=5)
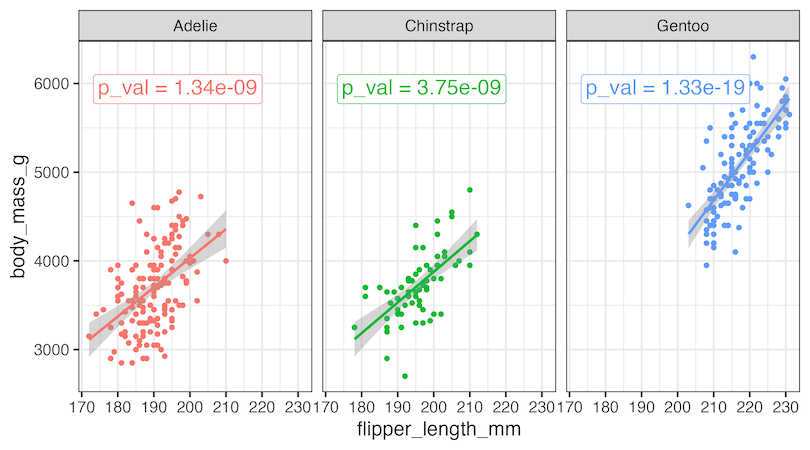
Annotating each facet with p-value using geom_richtext()
ggtext’s geom_richtext() function offers us great customizations for annotating plots. We can add matching colors and a box around p-value to highlight the annotation nicely.
p1+
geom_richtext(
data = pval_df,
aes(
label = label#,
),
hjust = 1, vjust = 1,
size=6,
font="bold"
)
ggsave("annotate_facet_wrap_plots_with_p_value_ggplot2.png", width=9, height=5)
If you want to annotate a single plot made with ggplot2, check out this post https://datavizpyr.com/annotate-plot-with-p-value-in-ggplot2/