In this post, we will learn how to visualize a dataframe with missing values represented as NAs as a heatmap. A quick visualization of missing values in the data is useful in analyzing the data. We will use mainly tidyverse approach, first to create a toy dataframe with missing values, then use ggplot2’s geom_tile() function to make the heatmap and add specific color to represent NAs using scale_fill_continuous() function.
Let us get started by loading tidyverse and palmer penguins dataset.
1 2 3 | library(tidyverse)library(palmerpenguins)theme_set(theme_bw(16)) |
We will consider only the numeric columns and select just the few rows for illustration.
1 2 3 4 5 | penguins <- penguins %>% select(where(is.numeric)) %>% select(-year) %>% drop_na() %>% head() |
Our toy data looks like this, six rows with no missing values.
1 2 3 4 5 6 7 8 9 10 11 | penguins# A tibble: 6 × 4 bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <dbl> <dbl> <int> <int>1 39.1 18.7 181 37502 39.5 17.4 186 38003 40.3 18 195 32504 36.7 19.3 193 34505 39.3 20.6 190 36506 38.9 17.8 181 3625 |
Adding NAs randomly to a dataframe using tidyverse
Let us add NAs to the toy dataframe randomly. We use across() function in combination with mutate() to introduce NAs in each column. We introduce NAs probabilistically with 20% chance for missing value at each element.
1 2 3 4 5 6 7 8 | set.seed(42)df <- penguins%>% mutate(across(where(is.numeric), ~ifelse(sample(c(TRUE, FALSE), size = n(), replace = TRUE, prob = c(0.8, 0.2)), ., NA))) |
Our data with missing values look like this.
1 2 3 4 5 6 7 8 9 10 11 | df # A tibble: 6 × 4 bill_length_mm bill_depth_mm flipper_length_mm body_mass_g <dbl> <dbl> <int> <int>1 NA 18.7 NA 37502 NA 17.4 186 38003 40.3 18 195 NA4 NA 19.3 NA 34505 39.3 20.6 NA NA6 38.9 17.8 181 NA |
Visualizing Dataframe with NAs as heatmap
To make a heatmap, we will use ggplot2’s geom_tile() function. First wee need to reshape the data into tidy long form using pivot_longer() function. We also add unique row number to help us make the heatmap.
1 2 3 4 5 | df_tidy <- df %>% scale() %>% as.data.frame() %>% mutate(row_id=row_number()) %>% pivot_longer(-row_id, names_to="feature", values_to="values") |
Our tidy data with NAs look like this
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | df_tidy# A tibble: 24 × 3 row_id feature values <int> <chr> <dbl> 1 1 bill_length_mm NA 2 1 bill_depth_mm 0.0566 3 1 flipper_length_mm NA 4 1 body_mass_g 0.440 5 2 bill_length_mm NA 6 2 bill_depth_mm -1.05 7 2 flipper_length_mm -0.188 8 2 body_mass_g 0.704 9 3 bill_length_mm 1.11 10 3 bill_depth_mm -0.538 # … with 14 more rows |
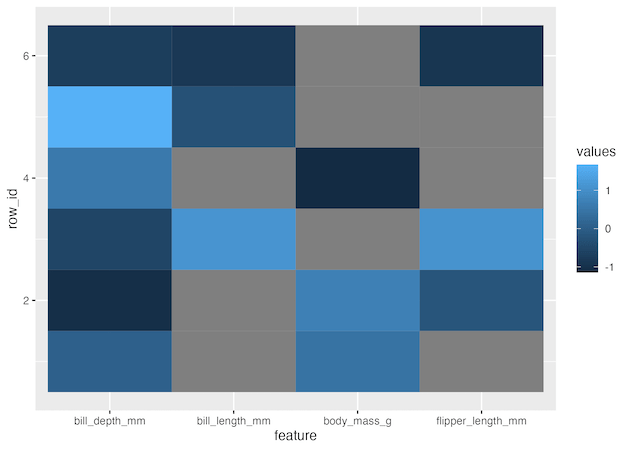
Now we can go ahead and make a heatmap using geom_tile() function. Here x-axis will be the penguin features and y axis is each penguin. We fill each tile by the values of the features. Note that we have scaled the values of each column as they were on very different scales.
1 2 3 | df_tidy %>% ggplot(aes(x=feature, y=row_id, fill=values))+ geom_tile() |
geom_tile() recognizes NA values in our data and colors them as grey.
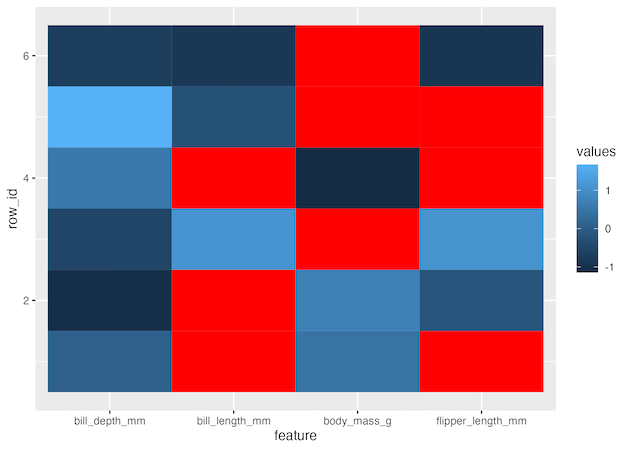
If needed we can customise the tiles for NAs with a color of choice using scale_fill_continuous().
1 2 3 4 | df_tidy %>% ggplot(aes(x=feature, y=row_id, fill=values))+ geom_tile()+ scale_fill_continuous(na.value = 'red') |
Also note that when we reshape our data from wide to long, the column order on the heatmap is different from the original column order. The heatmap’s column order is sorted alphabetically and it is the same order as shown below.
1 2 3 4 5 6 7 8 9 10 11 12 | df %>% select(sort(names(.)))# A tibble: 6 × 4 bill_depth_mm bill_length_mm body_mass_g flipper_length_mm <dbl> <dbl> <int> <int>1 18.7 NA 3750 NA2 17.4 NA 3800 1863 18 40.3 NA 1954 19.3 NA 3450 NA5 20.6 39.3 NA NA6 17.8 38.9 NA 181 |




