UMAP, short for “Uniform Manifold Approximation and Projection” is a one of the useful dimensionality reduction techniques like tSNE and PCA. UMAP is non-linear dimension reduction technique and often used for visualizing high-dimensional datasets. In this tutorial, we will learn how to perform dimensionality reduction using UMAP in R and learn make a UMAP plot using ggplot2 in R.
Loading Data and Packages
We will use Palmer Penguin dataset to make a UMAP plot in R. We will perform umap using the R package umap. Let us load the packages needed and set the simple b&w theme for ggplot2 using theme_set() function.
library(tidyverse)
library(palmerpenguins)
#install.packages("umap")
library(umap)
theme_set(theme_bw(18))
To perform UMAP using Palmer Penguin’s dataset, we will use numerical columns and ignore non-numerical columns as meta data (like we did it for doing tSNE analysis in R). First, let us remove any missing data and add unique row ID.
penguins <- penguins %>% drop_na() %>% select(-year)%>% mutate(ID=row_number())
## # A tibble: 6 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g sex ## <fct> <fct> <dbl> <dbl> <int> <int> <fct> ## 1 Adelie Torge… 39.1 18.7 181 3750 male ## 2 Adelie Torge… 39.5 17.4 186 3800 fema… ## 3 Adelie Torge… 40.3 18 195 3250 fema… ## 4 Adelie Torge… 36.7 19.3 193 3450 fema… ## 5 Adelie Torge… 39.3 20.6 190 3650 male ## 6 Adelie Torge… 38.9 17.8 181 3625 fema… ## # … with 1 more variable: ID <int>
Let us create a dataframe with all categorical variables with the unique row ID.
penguins_meta <- penguins %>% select(ID, species, island, sex)
Performing UMAP with umap package
Let us select numerical columns using is.numeric() function with select(), standardise the data using scale() function before applying umap() function to perform tSNE.
set.seed(142)
umap_fit <- penguins %>%
select(where(is.numeric)) %>%
column_to_rownames("ID") %>%
scale() %>%
umap()
The umap result object is a list object and the layout variable in the list contains two umap components that we are interested in. We can extract the components and save it in a dataframe. Also, we merge the UMAP components with the meta data associated with the data.
umap_df <- umap_fit$layout %>%
as.data.frame()%>%
rename(UMAP1="V1",
UMAP2="V2") %>%
mutate(ID=row_number())%>%
inner_join(penguins_meta, by="ID")
umap_df %>% head() ## UMAP1 UMAP2 ID species island sex ## 1 -7.949633 -1.387130 1 Adelie Torgersen male ## 2 -6.850185 -1.685802 2 Adelie Torgersen female ## 3 -6.753245 -2.485241 3 Adelie Torgersen female ## 4 -9.327034 -1.900235 4 Adelie Torgersen female ## 5 -10.353931 -1.381105 5 Adelie Torgersen male ## 6 -7.273715 -1.689724 6 Adelie Torgersen female
UMAP plot: Scatter plot between two UMAP components
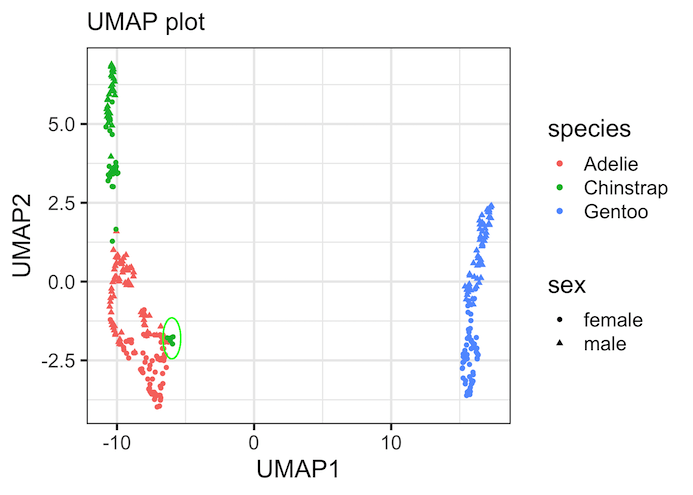
We can make UMAP plot, a scatter plot with the two UMAP components colored by variables of interest that are part of the data. In this example, we have added color by species variable and shape by sex variable.
umap_df %>%
ggplot(aes(x = UMAP1,
y = UMAP2,
color = species,
shape = sex))+
geom_point()+
labs(x = "UMAP1",
y = "UMAP2",
subtitle = "UMAP plot")
ggsave("UMAP_plot_example1.png")
Our UMAP plot looks like this. Note, UMAP is unsupervised technique and has nicely identified three groups corresponding the species variable in the data.

UMAP plot in R: Example 2
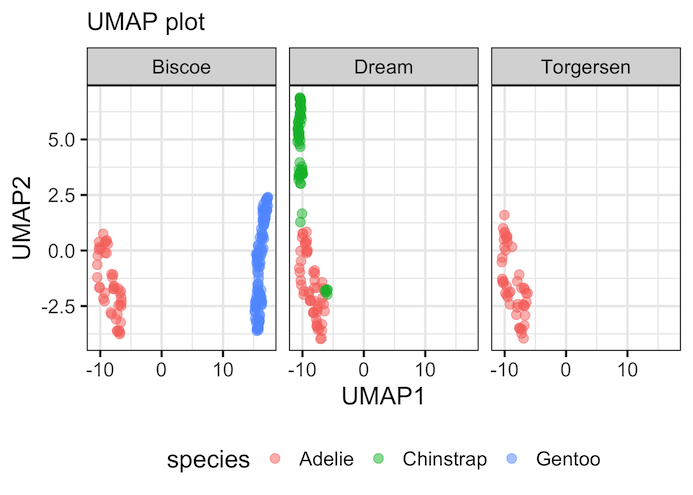
In the second example of UMAP plot, we have used the same UMAP components, but this time we have added facetting based on island variable to see the relationship between species and island more clearly.
umap_df %>%
ggplot(aes(x = UMAP1,
y = UMAP2,
color = species)) +
geom_point(size=3, alpha=0.5)+
facet_wrap(~island)+
labs(x = "UMAP1",
y = "UMAP2",
subtitle="UMAP plot")+
theme(legend.position="bottom")
ggsave("UMAP_plot_example2.png")
UMAP plot to identify potential sample mixup issue or outliers
One of the biggest advantages of unsupervised/dimensionality techniques like UMAP or tSNE is that it can detect patterns in the data and force us to rethink about the annotations of the dataset. For example, in this Palmer penguin data, UMAP plot shows a few Chinstrap Penguin samples (in green) are within the Adelie samples (in red). It suggests a possible sample annotation mix-ups or outliers.
library(ggforce)
umap_df %>%
ggplot(aes(x = UMAP1,
y = UMAP2,
color = species,
shape = sex)) +
geom_point() +
labs(x = "UMAP1",
y = "UMAP2",
subtitle="UMAP plot") +
geom_circle(aes(x0 = -6, y0 = -1.8, r = 0.65),
color = "green",
inherit.aes = FALSE)
ggsave("umap_plot_to_identify_outlier_samples.png")